|
This site describes the set up and usage of `comet', an actin-based bead motility simulator.
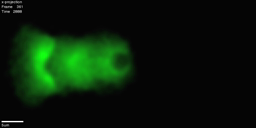
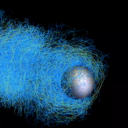
Actin is a polymer built by cells that allows them to move. Actin-based bead motility is an in vitro model system used to study how our cells produce force and move, carried out by coating a bead with proteins that tell the cell to polymerize actin, and putting the bead a solution that mimics the material inside cells. This causes an actin network to build around the bead. Surprisingly even when the bead is spherically symmetric, rather than just building a symmetric shell that gets bigger and bigger, it moves off in 'comet tail' of actin. This bead motility simulator aims to help us understand how this process works.
Simulator Output
Example Results
Model Robustness
In Vitro
Essential Information
Installing the program
comet runs on any unix-like operating system, including OS X, Linux or Windows (under cygwin).
Running the program
The program is called from the command line. The command line parameters tell the program what to do (calculate a new run, re-process existing data, interactive 3D view etc.). A cometparams.ini file in the working directory tells the program detailed information about the model parameters to use for calculation and display.
How the program works
In Depth
|
Code Status
|
|